Introduction
Many people have asked me this question that whenever they get started with data science, they get stuck with the manifold of tools available to them.
Although there are handful of guides available out there concerning the problem such as “19 Data Science Tools for people who aren’t so good at Programming” or “A Complete Tutorial to Learn Data Science with Python from Scratch“, I would like to show what tools I generally prefer for my day-to-day data science needs.
Read on if you are interested!
Note: I usually work in Python and in this article I intend to cover the tools used in python ecosystem on Windows.
Table of Contents
- What does a “data science stack” look like?
- Case Study of a Deep Learning problem: Getting started with Python Ecosystem
- Overview of Jupyter: A Tool for Rapid Prototyping
- Keeping Tab of Experiments: Version Control with GitHub
- Running Multiple Experiments at the Same Time: Overview Tmux
- Deploying the Solution: Using Docker to Minimize Dependencies
- Summarization of tools
What does a “data science stack” look like?
I come from a software engineering background. There people are inquisitive about what kind of “stack” people currently working on? Their intention here is to find which tools are trending and what are the pros and cons while using it. Its because each and everyday new tools come out which try to eliminate the nagging problems people face everyday.
In data science, I can probably say that there is more inclination on what kinds of techniques you use to solve a problem rather than the tools to use. Still, its wise to get to know what kind of tools are available to you. A survey was done keeping this in mind. Below image summarizes these findings.
Case Study of a Deep Learning problem: Getting started with Python Ecosystem
Instead of blatantly saying which the tools to be used, I will give you a rundown of these tools with a practical example. We will do this exercise on “Identify the Digits” practice problem.
Let us first get to know what the problem entails. The home page mentions “Here, we need to identify the digit in given images. We have total 70,000 images, out of which 49,000 are part of train images with the label of digit and rest 21,000 images are unlabeled (known as test images). Now, we need to identify the digit for test images.”
This essentially means that the problem is an image recognition problem.The first step here would be to setup your system for the problem. I usually create a specific folder structure for each problem I start (yes I’m a windows user 😐 ) and start working from there.
For this kind of problems, I have a tendency to use the kit mentioned below:
- Python 3 to get started of course!
- Numpy / Scipy for basic data reading and processing
- Pandas to structure the data and bring it in shape for processing
- Matplotlib for data visualization
- Scikit-learn / Tensorflow / Keras for predictive modelling
Fortunately, most of the things above can be accessed using a single software called Anaconda. I’m accustomed to using Anaconda because of its comprehensiveness of data science packages and ease of use.
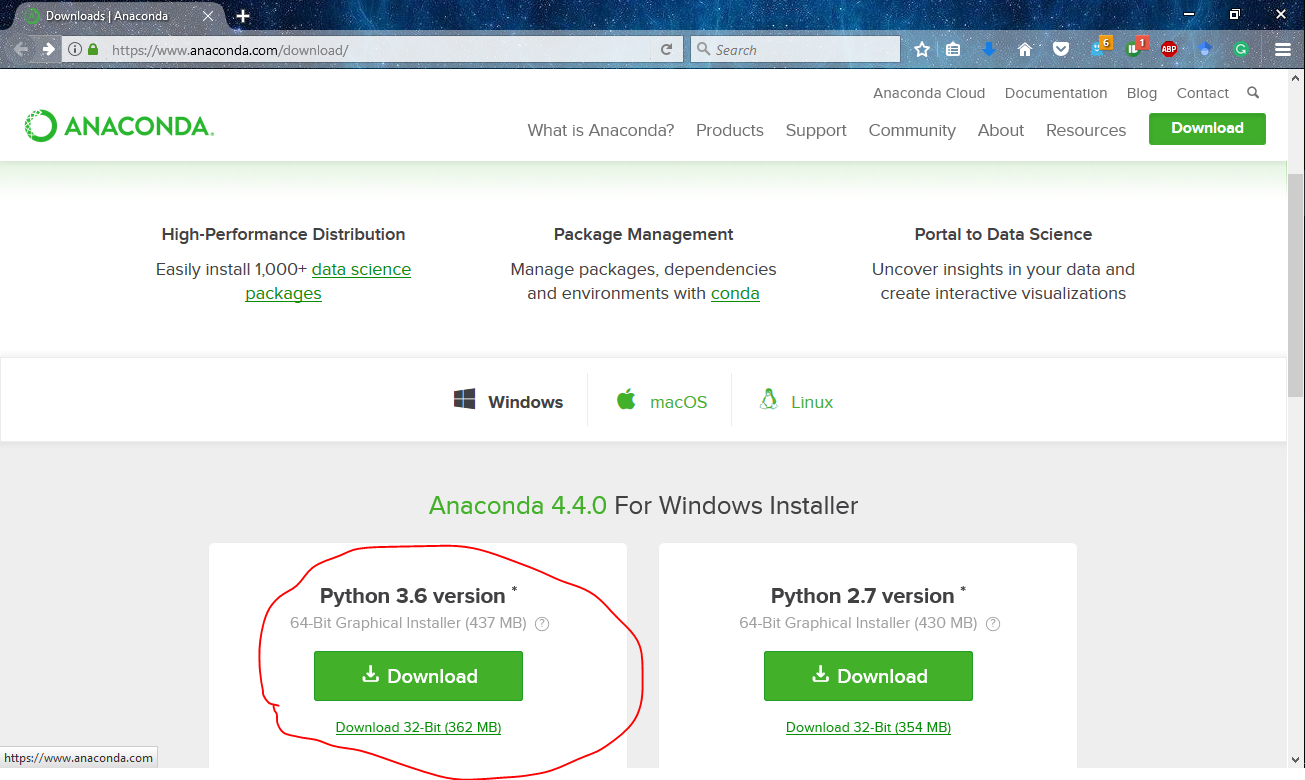
To setup anaconda in your system, you have to simply download the appropriate version for your platform. More specifically, I have the python 3.6 version of anaconda 4.4.0.
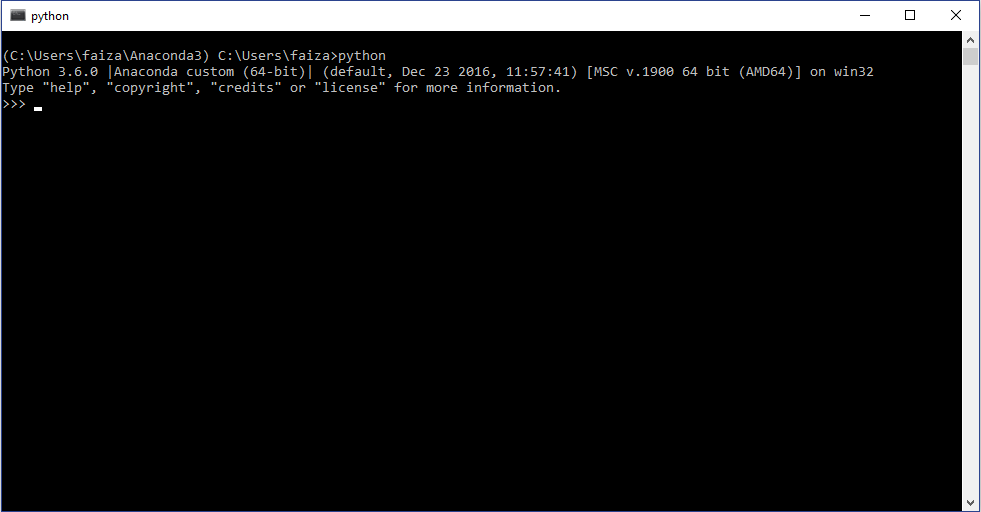
Now to use the newly install python ecosystem, open the anaconda command prompt and type “python”
As I said earlier,most of the things come pre-installed in anaconda. The only libraries left are tensorflow and keras. A smart thing to do here which anaconda provides a feature for is creating an environment. You do this because even if you do something wrong when setting up, it won’t affect your original system. This is like creating a sandbox for all your experiments. To do this, go to the anaconda command prompt and type
conda create -n dl anaconda
Now not install , the remaining packages by typing
pip install --ignore-installed --upgrade tensorflow keras
Now you can start writing your codes in your favorite text editor and run the python scripts!
Overview of Jupyter: A Tool for Rapid Prototyping
The problem in working with a plain text editor is that each time you update something, you have to run the code from the start again. Suppose you have a small code for reading data and processing. The code for data reading is functioning correctly but takes an hour to run. Now if you try to change the code for processing, you have to wait for the code for data reading to run and then see if your update works. This is tiresome and it wastes your time a lot.
To get over this issue, you can use jupyter notebooks. Jupyter notebooks essentially save your progress and let you continue from where you left off. Here you can write your code in a structured way so that you can resume the code and update it whenever you want to.
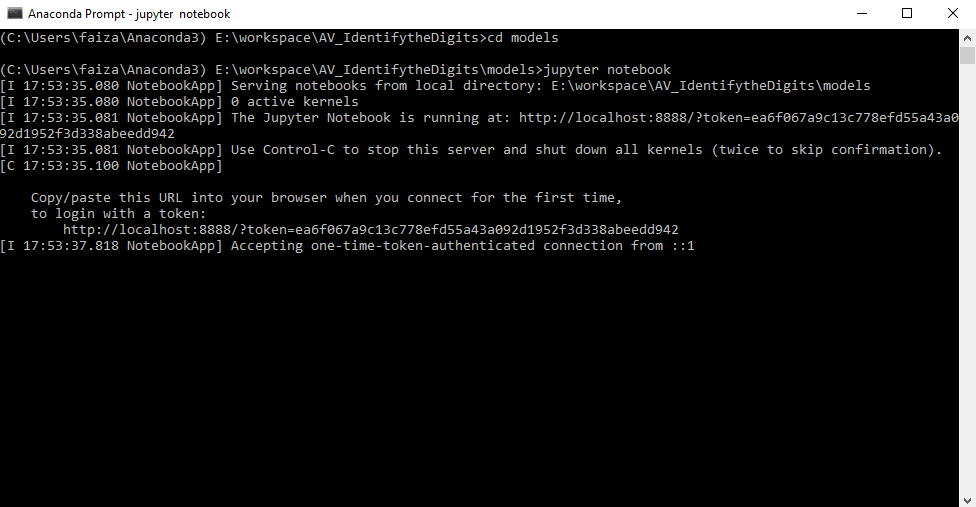
In the section above, you had setup your system with anaconda software. As I mentioned, anaconda has jupyter preinstalled in it. To open jupyter notebook, open the anaconda prompt, go to the directory that you created and type
jupyter notebook
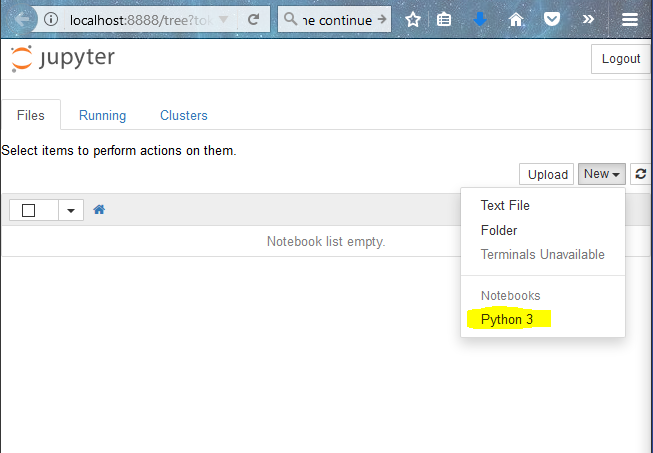
This opens up jupyter notebook in you web browser (Mozilla Firefox for me). Now to create a notebook, click on “New”-> “Python 3”
Now what i usually do is divide the code into small blocks of code, so that it would be easier for me to debug. I always keep these in mind when I write code:
- Note down in plain english what I want to do
- Import all the necessary libraries required
- Write as much generic code as possible so that I would not have to update much.
- Test every little thing as and when I do it.
Here is a sample of code I wrote to solve the deep learning problem mentioned. You can follow along if you like!
Keeping Tab of Experiments: Version Control with GitHub
A data science project requires multiple iterations of experimentation and testing. It is very difficult to remember all the things you tried out, which one of those worked and which did not.
One method to control this issue is to take notes of all the experiments that you did and summarize them. But this too is a bit tedious and manual work. A workaround for this (and my personal choice) is to use GitHub. Originally GitHub was used as a code sharing platform for software engineers, but now it is gradually being accepted by the data science community. What GitHub does is provides you with a framework for saving all the changes you did in the code and reverting back to it anytime you want. This gives you the flexibility that you need to do a data science project efficiently.
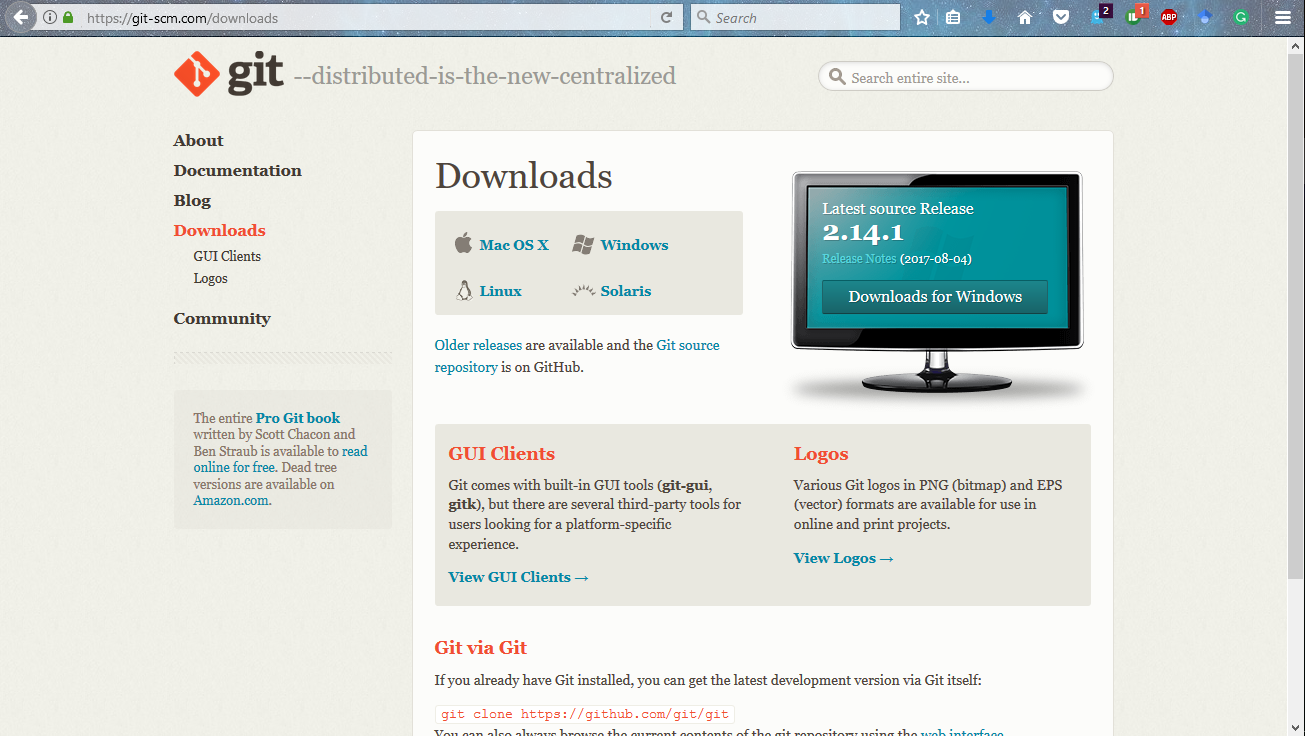
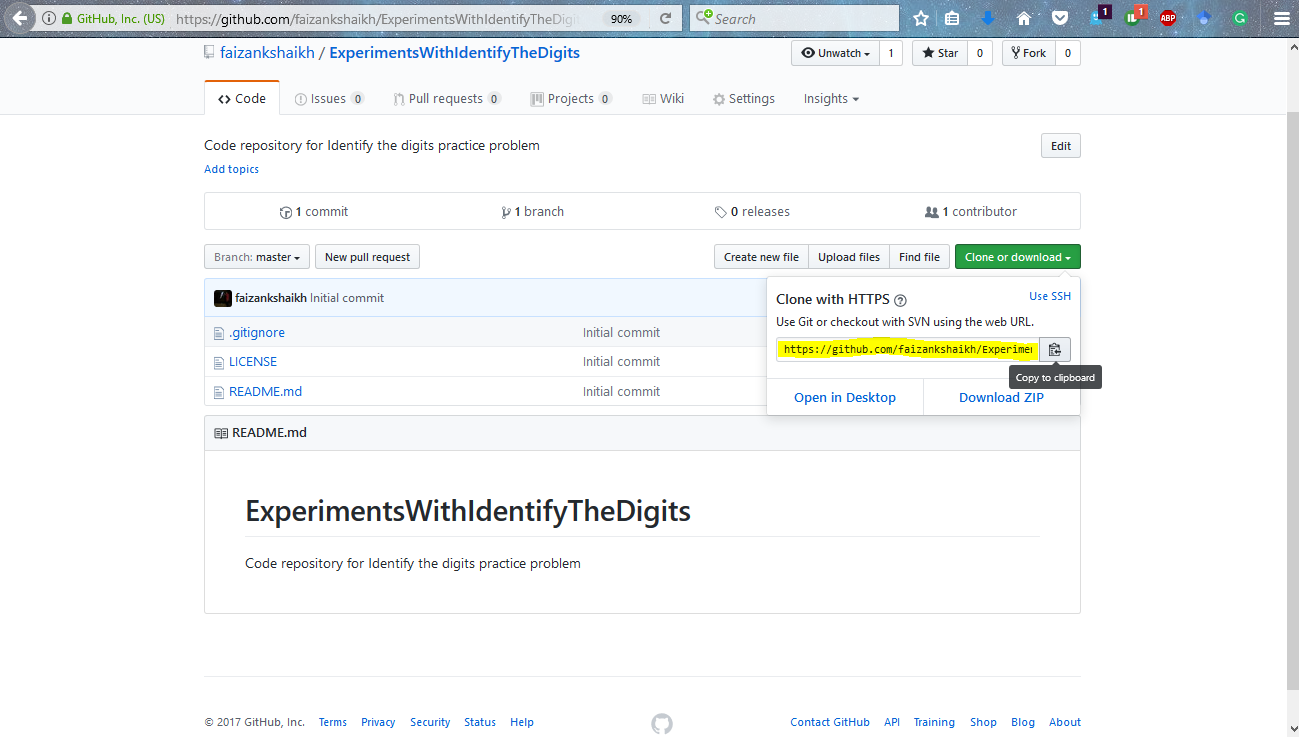
I will give you an example of how to use GitHub. But first, let us install it in our system. Go to the download link and click on the version as per your system. Let it download and then install it.
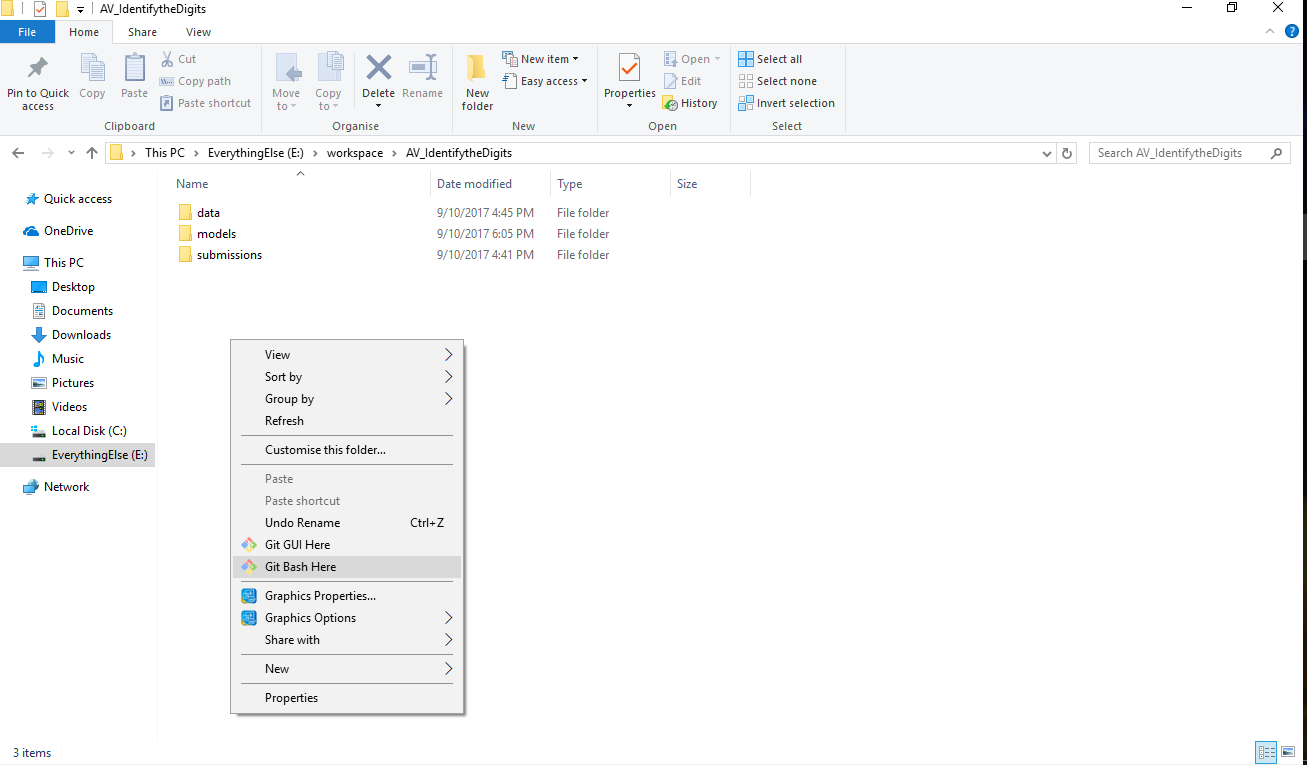
This will install git in your system, along with command prompt for it called git bash. The next step is to configure the system with your git account. Once you have signed up for GitHub, you can use this to setup your system. Open up you git bash and type the following commands.
git config --global user.name "jalFaizy"
git config --global user.email [email protected]Now there’s two tasks you have to do.
- Create a repository on GitHub
- Connect it with your code directory.
To accomplish the tasks, follow on to the steps mentioned below
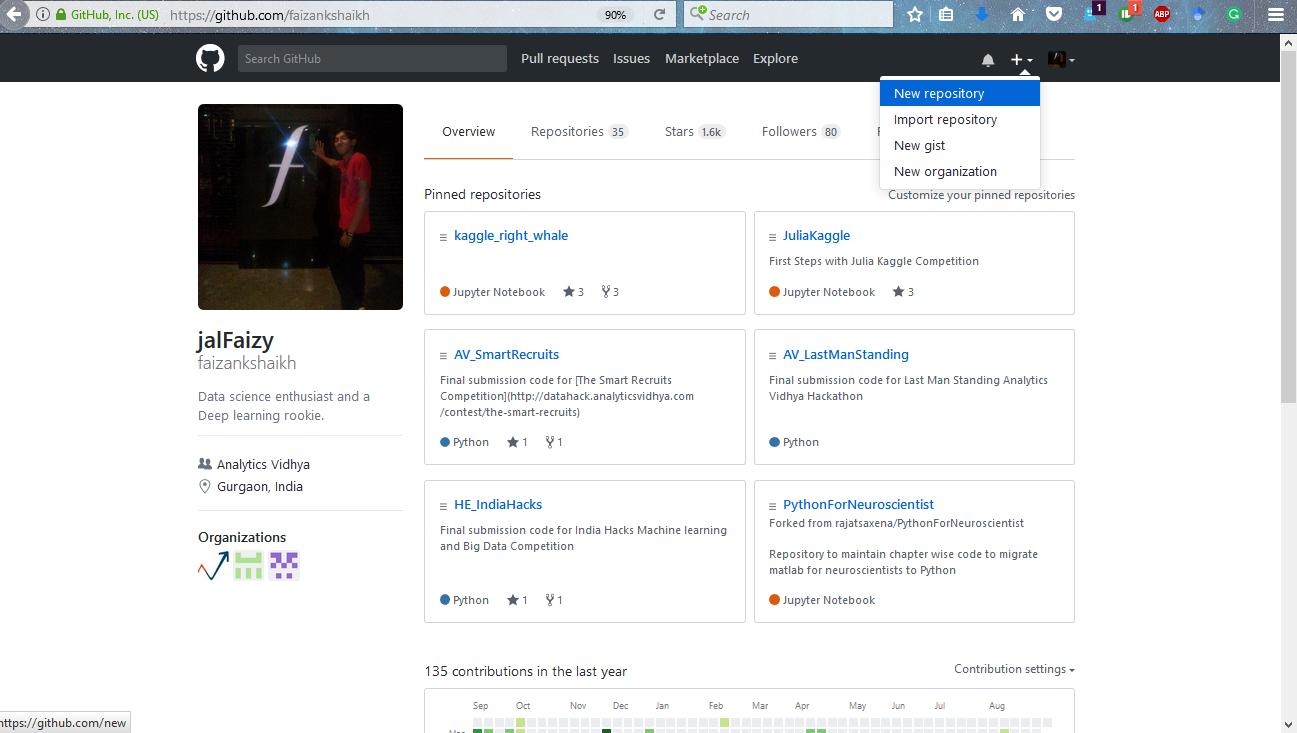
Step 1: Add a new repository on GitHub
Step 2: Give proper descriptions to setup the repository.
Step 4: Connect you local repository with the repository on GitHub with the commands below.
git init git remote add origin <add link here> git pull origin master
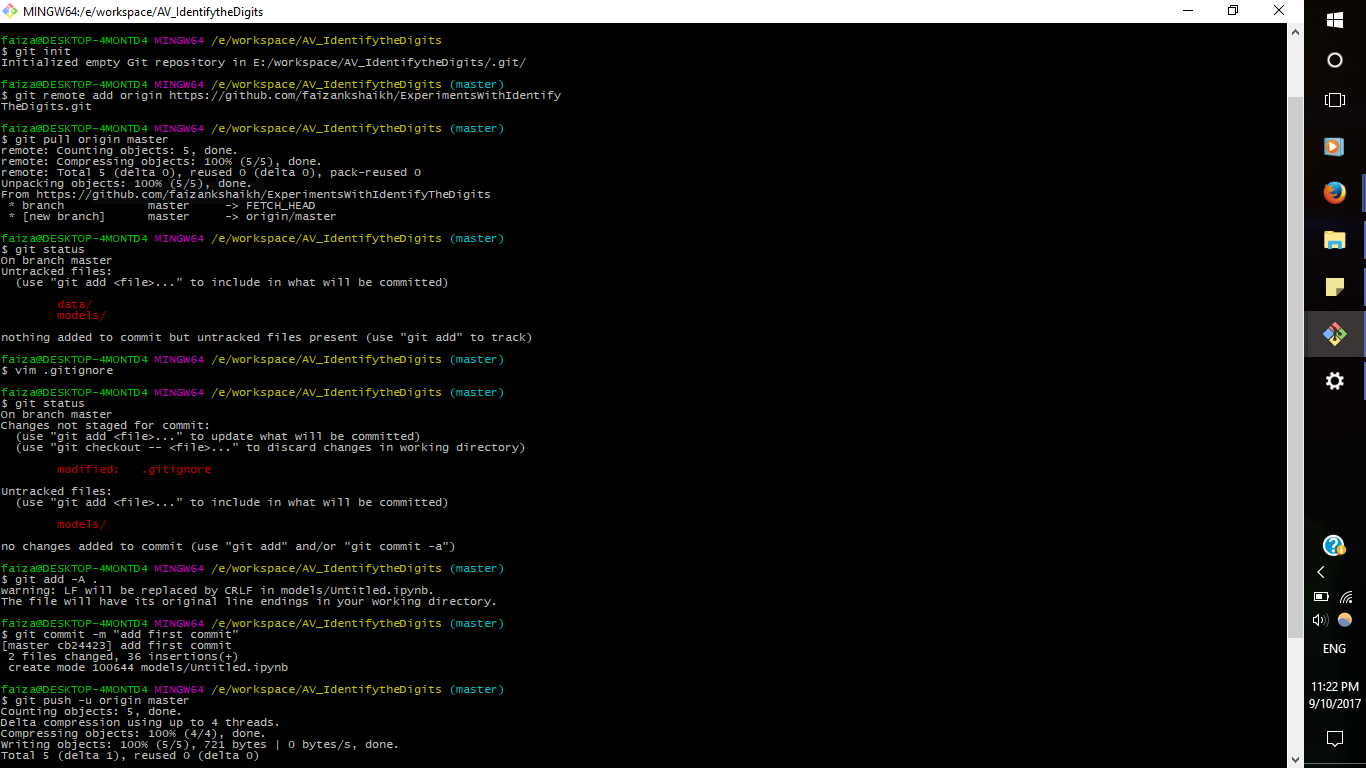
Now if you want GitHub to ignore the changes in the files you can mention them in .gitignore file. Open it and add the file or folder you want to ignore.
git add -A . git commit -m "add first commit" git push -u origin master
Now each time you want to save your progress, run these commands again
git add -A . git commit -m "<add your message>" git push -u origin master
This will ensure that you can go back to where you left off!
Running Multiple Experiments at the Same Time: Overview Tmux
Jupyter notebooks are very helpful for our experimentations. But what if you wanted to multiple experiments at the same time? You would have to wait for the previous command to finish before you can run another one.
I usual have many ideas that I want to test out. And there are times I want to scale up. I use the company’s deep learning box we built a few months back. So I prefer to use tmux for it. tmux or Terminal Multiplexer lets you switch easily between several programs in the same terminal. Here is my command center right now 🙂
We setup Ubuntu in the monster because it seemed like a good idea at that time. Setting up tmux in your system is pretty easy, you just have to install it using the command below
sudo apt-get install tmux
To open tmux, type “tmux” in your command line.
To make a new window, just press control + B and then shift + 5
Now if you want to let this experiment continue and do something else, just type
tmux detach
and it will give you the terminal again. And if you want to come back to the experiment again, type
tmux attach
As simple as that! Note that setting up tmux in windows is a bit different. You can refer this article to set it up in your system.
Deploying the Solution: Using Docker to Minimize Dependencies
Now after all the implementations are done, we still have to deploy the solutions so that the end user such as a developer can access it. But the issue we always face is that the system we have might not be the same as that of the user. There will always be installation and setting up issues in their system.
This is a very big problem when it comes to deployment of products in market. So to curb this issue, you can rely on a tool called docker. Docker is works on the idea that you can package code along with its dependencies into a self-contained unit. This unit can then be distributed to the end user.
I usually do toy problems on my local machine, but when it comes to final solutions, I rely on the monster. I had setup docker in that system using these commands
# docker installation
sudo apt-get update
sudo apt-get install apt-transport-https ca-certificates
sudo apt-key adv \
--keyserver hkp://ha.pool.sks-keyservers.net:80 \
--recv-keys 58118E89F3A912897C070ADBF76221572C52609D
echo "deb https://apt.dockerproject.org/repo ubuntu-xenial main" | sudo tee /etc/apt/sources.list.d/docker.list
sudo apt-get update
sudo apt-get install linux-image-extra-$(uname -r) linux-image-extra-virtual
sudo apt-get install docker-engine
sudo service docker start
# setup docker for another user
sudo groupadd docker
sudo gpasswd -a ${USER} docker
sudo service docker restart
# check installation
docker run hello-world
Integrating GPU on docker has some additional steps:
wget -P /tmp https://github.com/NVIDIA/nvidia-docker/releases/download/v1.0.0-rc.3/nvidia-docker_1.0.0.rc.3-1_amd64.deb sudo dpkg -i /tmp/nvidia-docker*.deb && rm /tmp/nvidia-docker*.deb # Test nvidia-smi nvidia-docker run --rm nvidia/cuda nvidia-smi
Now comes the main part, installing DL libraries on docker. We built the docker from scratch, following this excellent guide (https://github.com/saiprashanths/dl-docker/blob/master/README.md)
git clone https://github.com/saiprashanths/dl-docker.git cd dl-docker docker build -t floydhub/dl-docker:gpu -f Dockerfile.gpu
To run your code on docker, open your docker’s command prompt using the command
nvidia-docker run -it -p 8888:8888 -p 6006:6006 floydhub/dl-docker:gpu bash
Now whenever I have a complete working model, I throw it in the docker system to try it out on scale. You can use the dockerfile in a different system to install the same softwares and libraries in their system. This ensures that there isn’t any installation problem.
Summarization of tools
To conclude these are the tools I use and their usages
- Anaconda for setting up the python ecosystem and the essential packages
- Jupyter Notebooks for continuous experimentation
- GitHub to save my progress of experiments
- Tmux for running multiple experiments at once
- and lastly, docker for deployment
Hope these tools will help you in your experiments too!
End Notes
In this article, I have covered what I feel are the necessarily tools for doing a data science project. To summarize, I use Python environment in anaconda stack, Jupyter notebooks for experimentations, Github for saving the crucial experiments, Tmux for running multiple experiments at once, and docker for deployment.
If you have any more tools to suggest or if you use a completely different stack, do let me know in the comments below!