Learning new tools and techniques in data science is sort of like running on treadmill – you have to run continuously to stay on top of it. The minute you stop, you start falling behind.
As part of this learning, I continuously look out for new developments happening in new tools and techniques. It was in this desire to continuously learn that I came across Julia about a year back. It was in very early stages then – it still is!
But, there is something special about Julia, which makes it a compelling tool to learn for all future data scientists. So, I thought to write a few articles on it. This is first of these articles, which provides the motivation to learn Julia, its installation, current packages available and ways to become part of Julia community.
What is Julia?
Julia is a high-level, high-performance dynamic programming language for technical computing, with easy to write syntax. It provides a sophisticated compiler, distributed parallel execution, numerical accuracy, and an extensive mathematical function library.
Why another programming language?
The simplest way to understand its power is to think of it as a language which has a wide range of statistical packages like R, it is easy to write and learn like Python and has execution speed similar to C / C++. If you are still not convinced about what I have mentioned, have a look at benchmarks of a few common benchmarks below:
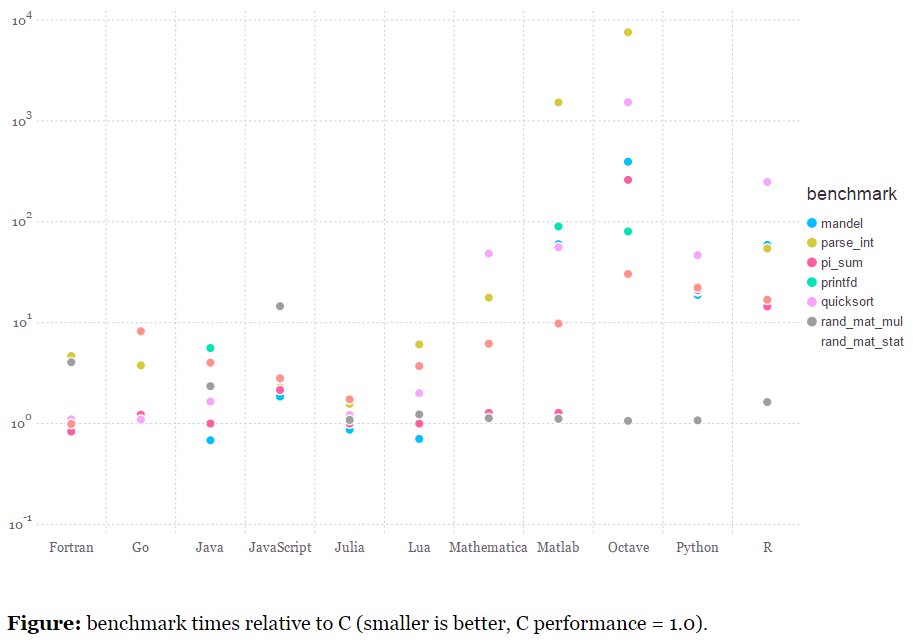
C compiled by gcc 4.8.2, taking best timing from all optimization levels (-O0 through -O3). C, Fortran and Julia use OpenBLAS v0.2.12. The Python implementations of rand_mat_stat and rand_mat_mul use NumPy (v1.8.2) functions; the rest are pure Python implementations.
A Summary of Features in Julia
Some of the important features to highlight from data science capabilities are:
- Good performance, approaching that of statically-compiled languages like C
- Built-in package manager to make life easier
- Lisp-like macros and other metaprogramming facilities
- Call C functions directly & Python functions: use the PyCall package
- Powerful shell-like capabilities for managing other processes
- Designed for parallelism and distributed computation
- User-defined types are as fast and compact as built-ins
- Automatic generation of efficient, specialized code for different argument types
- Elegant and extensible conversions and promotions for numeric and other types
- MIT licensed: free and open source
A more comprehensive list of features can be accessed here
Installation of Julia
Now that you might be raring to give Julia a try for all the promises made above, let me quickly walk through various options to test drive your new sedan (which has sports car like acceleration):
- Option 1: Try Juliabox in browser – The simplest of option – no setup required. Just go to Juliabox, sign in using Google (sorry, if you don’t have a Google account – try the next version) and your instance is ready to fire.
- Option 2 – Use an IDE – Juno seems to be the best IDE available right now. Sadly, JuliaStudio is no longer supported. The best way to install it is to download the combo package from Julia site itself.
- Option 3 – Using Command line – If you are the hardcore programmer, who can’t think of a programming language without a command line, don’t worry! There is an option for you as well. You can download the package here.
- Option 4 – Using iJulia notebooks – If you are a Python explorer and have used iPython for your interactive data exploration – here is an awesome news. iJulia notebooks are equally awesome and carry over similar interface. In order to install iJulia, you need to install iPython first, then install Julia 0.3 or later. Next start Julia and add package “IJulia” and start using it. You can find more details here.
The installation was pretty simple and straight forward. I have tried Juliabox as well as Juno. Option 1 and 2 come with a few demo examples before hand. You can just follow the comments (starting with #) to understand and give the code a test run.
A few important packages
There are a total of 610 packages on Julia as on date (9th July 2015). If you filter out packages for which tests have failed or which have not been tested, you are only left with 381 packages. Among these I have filtered out the ones related to data science and have more than 15 stars. That leaves us with the following packages:
| Package | Description | Version | Stars |
| BackpropNeuralNet | A neural network in Julia | 0.0.3 | 18 |
| Bokeh | Bokeh Bindings for Julia | 0.1.0 | 26 |
| Boltzmann | Restricted Boltzmann Machines in Julia | 0.1.0 | 19 |
| Calculus | Calculus functions in Julia | 0.1.8 | 46 |
| Clustering | A Julia package for data clustering | 0.4.0 | 33 |
| Convex | A julia package for disciplined convex programming. | 0.0.6 | 108 |
| Cpp | Utilities for calling C++ from Julia | 0.1.0 | 18 |
| DataArrays | Data structures that allow missing values | 0.2.16 | 21 |
| DataFrames | library for working with tabular data in Julia | 0.6.7 | 206 |
| DataFramesMeta | Metaprogramming tools for DataFrames | 0.0.1 | 33 |
| DataStructures | Julia implementation of Data structures | 0.3.10 | 52 |
| DecisionTree | Decision Tree Classifier and Regressor | 0.3.8 | 36 |
| Distances | A package for evaluating distances(metrics) between vectors. | 0.2.0 | 21 |
| Distributions | A package for probability distributions & associated functions. | 0.7.4 | 101 |
| DSP | Filter design, periodograms, window functions, and other digital signal processing functionality | 0.0.8 | 32 |
| FunctionalCollections | Functional and and persistent data structures for Julia | 0.1.2 | 34 |
| Gadfly | Crafty statistical graphics for Julia. | 0.3.13 | 684 |
| GeneticAlgorithms | A lightweight framework for writing genetic algorithms in Julia | 0.0.3 | 86 |
| GLM | Generalized linear models in Julia | 0.4.6 | 78 |
| GLMNet | Wrapper for fitting Lasso/ElasticNet GLM models using glmnet | 0.0.4 | 23 |
| Graphs | Working with graphs in Julia | 0.5.5 | 90 |
| HDF5 | Saving and loading Julia variables | 0.4.18 | 65 |
| HypothesisTests | Hypothesis tests for Julia | 0.2.9 | 16 |
| Images | An image library for Julia | 0.4.39 | 73 |
| JuMP | Modeling language for Mathematical Programming (linear, mixed-integer, conic, nonlinear) | 0.9.2 | 162 |
| MachineLearning | Julia Machine Learning library | 0.0.3 | 37 |
| Mamba | Markov chain Monte Carlo (MCMC) for Bayesian analysis in julia | 0.4.11 | 44 |
| Markdown | Markdown parsing for Julia | 0.3.0 | 21 |
| Match | Advanced Pattern Matching for Julia | 0.1.3 | 29 |
| MixedModels | A Julia package for fitting (statistical) mixed-effects models | 0.3.22 | 41 |
| MLBase | A set of functions to support the development of machine learning algorithms | 0.5.1 | 41 |
| Mocha | Deep Learning framework for Julia | 0.0.8 | 297 |
| MultivariateStats | A Julia package for multivariate statistics & data analysis (e.g. dimension reduction) | 0.2.1 | 21 |
| NLopt | Package to call the NLopt nonlinear-optimization library from the Julia language | 0.2.1 | 31 |
| OpenStreetMap | Julia OpenStreetMap Package | 0.8.1 | 20 |
| Optim | Optimization functions for Julia | 0.4.2 | 116 |
| Orchestra | Heterogeneous ensemble learning for Julia. | 0.0.5 | 27 |
| PGM | A Julia framework for probabilistic graphical models. | 0.0.1 | 25 |
| PyCall | Package to call Python functions from the Julia language | 0.8.1 | 183 |
| RCall | Embedded R within Julia | 0.2.1 | 16 |
| RDatasets | Julia package for loading many of the data sets available in R | 0.1.2 | 34 |
| Regression | Algorithms for regression (e.g. linear / logistic regression) | 0.3.2 | 17 |
| Rif | Julia-to-R interface | 0.0.12 | 47 |
| StatsBase | Basic statistics for Julia | 0.6.15 | 57 |
| StreamStats | Compute statistics over data streams in pure Julia | 0.0.2 | 27 |
| TimeSeries | Time series toolkit for Julia | 0.5.10 | 37 |
P.S. There is a lot of development happening on the language and the libraries. So this can change very quickly.
A few things to note:
- Gadfly looks to be the most popular package. This might well be because it is being used as a showcase library across all the products in the ecosystem
- The core data science libraries look more evolved than some of the other libraries. Mocha for DeepLearning, Orchestra for optimization, DataFrames or distributions are all on more evolved version comparatively
How to install & use a package?
Installing and using a package in Julia is dead simple. If you want to install / add a package, simply type this in your programming interface
Pkg.add("Gadfly")
This will install the package as well as its dependencies.
Once the package is installed, you can load it simply by calling “using”
using Gadfly
Simple!
The Julia ecosystem:
Julia is supported by a close knit community of developers. Here are a few mailing lists, you can be a part of:
- julia-news – for important announcements, such as new releases.
- julia-users – discussion around the usage of Julia. New users of Julia can ask their questions here.
- julia-stats – special purpose mailing list for discussions related to statistical programming with Julia. Topics of interest include DataFrame support, GLM modeling, and automatic generation of MCMC code for Bayesian models.
- julia-opt – discussions related to numerical optimization in julia. This includes Mathematical Programming (linear, mixed-integer, conic, semi-definite, etc.), constrained and unconstrained gradient-based and gradient-free optimization, and related topics.
In addition to these newsletter, you can also look at juliabloggers.com . The site looks like a developing ecosystem as of now though.
End Notes
I hope that you have got a good overview of this powerful language under development. I was pretty excited when I saw it first and I continue to check this language for new developments closely. In the next articles to come, we will understand the data structured available in Julia, its interface with other languages e.g. Python and solve one of the case studies using Julia to understand its power.
What do you think of Julia? Are you all set to give it a try? Does the future excite you? Do let us know your thoughts through comments below.