This article was published as a part of the Data Science Blogathon
Research is to see what everybody else has seen and to think what
nobody else has thought – Albert Szent-Gyorgyi
Introduction:
As the business has realized the importance of data and the benefits of its right usage, the size of the data science teams has increased over the years. More teams are carrying out various experiments, revisions, and optimizations. It can become very complex in no time if a process is not brought in the place where every experiment is tracked, measured and results documented for reference. This goes a long way in avoiding redundant research and experiments.
To achieve this, replicability and reproducibility place an important role i.e is the ability to perform data analysis and achieve the same results as someone else.
Why do we need reproducible reports?

In this article, we will explore the process of building and managing machine learning reports using configuration files and generate HTML reports. For this simple machine learning project, I will use the Breast Cancer Wisconsin (Diagnostic) Data Set. The objective of this ML project is to predict whether a person has a benign or malignant tumour.
Let’s get started !!
- We will first conventionally build a classification model.
- We will build the same model using the YAML configuration file.
- Finally, we will generate an HTML report and save it.
Classification model – without config file:
Let’s create a Jupyter notebook by name notebook.ipynb and have the below code in it. I am using VSCode as my editor, it gives a nice and easier way to create a Jupyter notebook.
#mport important packages
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
import joblib
#path to the dataset
#filename = "../Data/breast-cancer-wisconsin.data"
filename = "./Data/breast-cancer-wisconsin.csv"
#load data
data = pd.read_csv(filename)
#replace "?" with -99999
data = data.replace('?', -99999)
# drop id column
data = data.drop(['id'], axis=1)
# Define X (independent variables) and y (target variable)
X = data.drop(['diagnosis','Unnamed: 32'], axis=1)
y = data['diagnosis']
#split data into train and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# call our classifer and fit to our data
classifier = KNeighborsClassifier(n_neighbors=5, weights="uniform",
algorithm = "auto", leaf_size = 25,
p=1, metric="minkowski", n_jobs=-1)
#training the classifier
classifier.fit(X_train, y_train)
#test our classifier
result = classifier.score(X_test, y_test)
print("Accuracy score is. {:.1f}".format(result))
#save our classifier in the model directory
joblib.dump(classifier, './Model/knn.pkl')
If you notice, in the above code there are various hardcoded numbers, file names, model parameters, train/test split percentage, etc. If you wish to experiment then you can make changes in the code and re-run it.
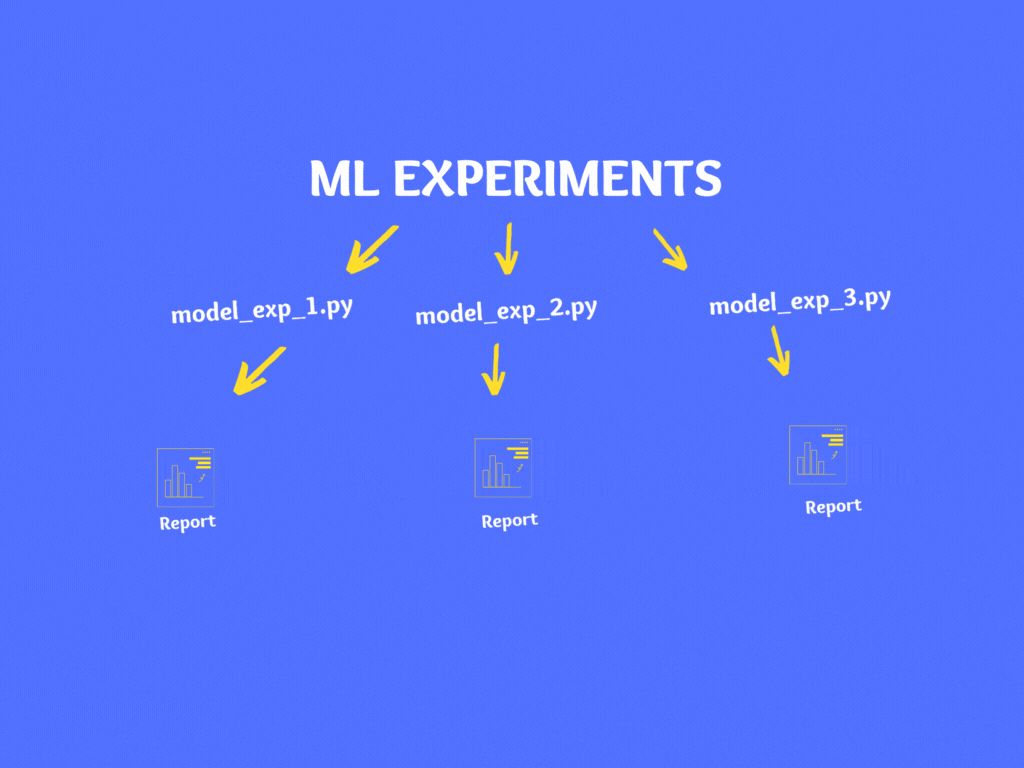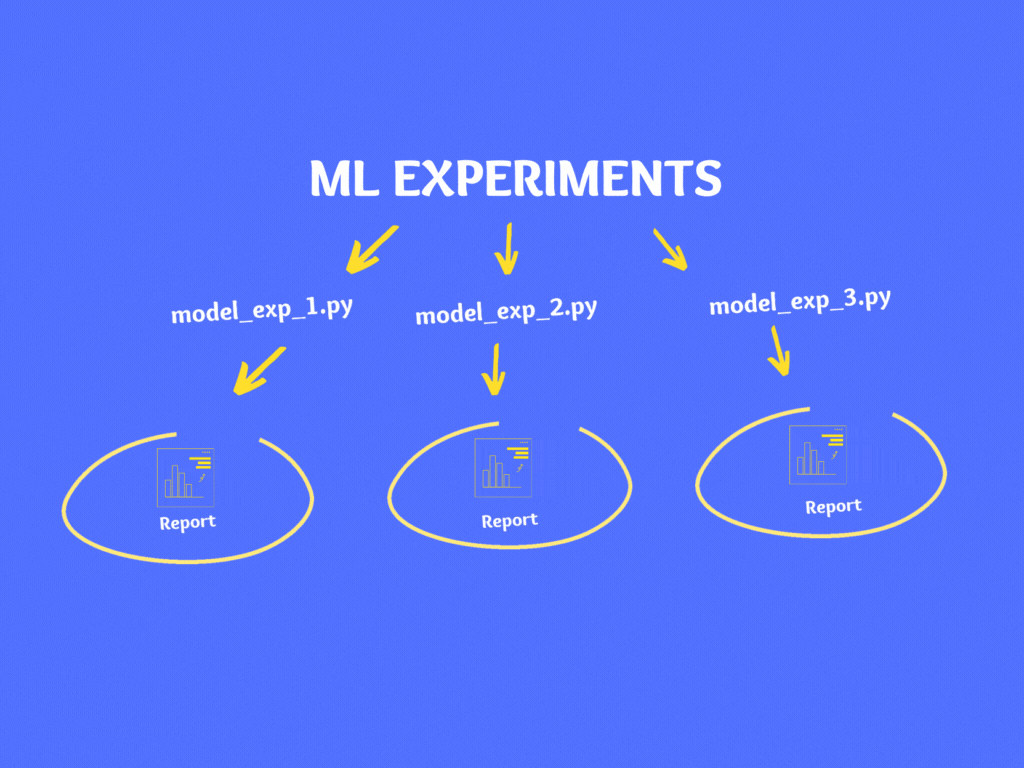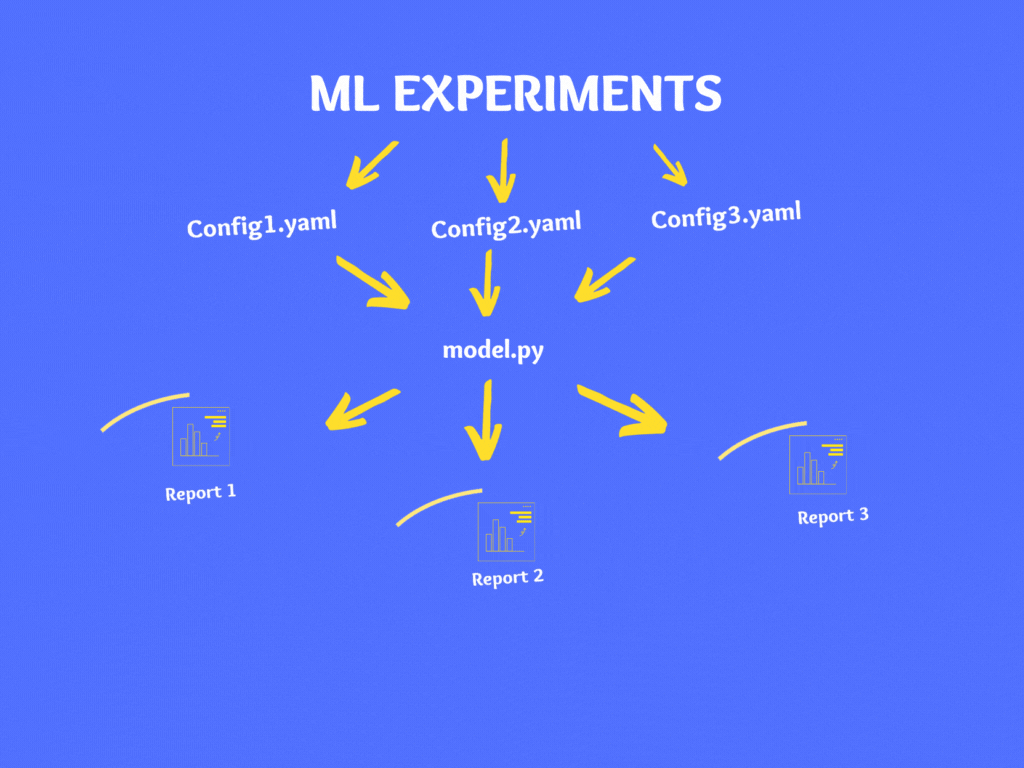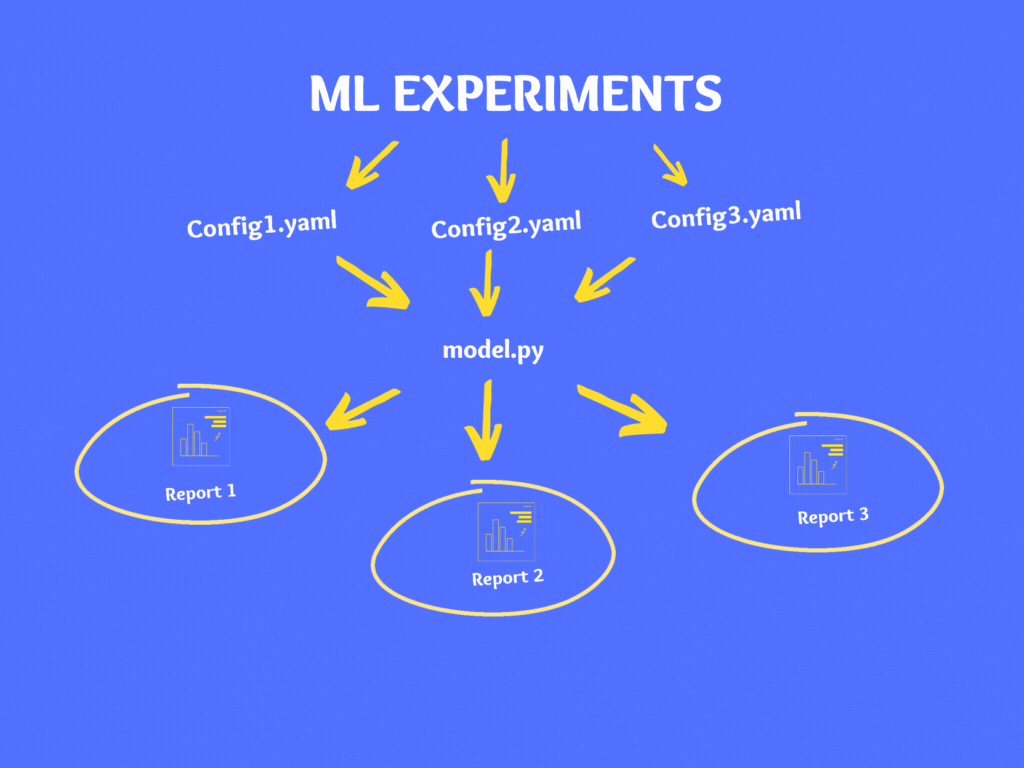
As a best practice, it is not advisable to make changes to code directly instead it is recommended to use configuration files. There are various file types for configuration like YAML, JSON, XML, INI, etc. and in our case, we will use the YAML format.
YAML file formats are popular for their ease of readability. YAML is relatively easy to write and within simple YAML files, there are no data formatting items, such as braces and square brackets; most of the relations between items are defined using indentation.
Let’s create our config file in YAML – Config.YAML
#INITIAL SETTINGS data_directory: "./Data/" data_name: "breast-cancer-wisconsin.csv" drop_columns: ["id","Unnamed: 32"] target_name: "diagnosis" test_size: 0.3 random_state: 123 model_directory: "./Model" model_name: KNN_classifier.pkl #kNN parameters n_neighbors: 3 weights: uniform algorithm: auto leaf_size: 15 p: 2 metric: minkowski n_jobs: 1
Now that we have built our model the conventional way, let’s move to the next section where we will do it slightly differently.
Classification model – with a config file:
There are two major changes compared to the last approach.
- Loading and reading of the YAML file.
- Replacing all the hardcoded parameters with variables from the YAML config file.
Let’s the below chunk of code to notebook.ipynb which will load the Config.yaml.
# folder to load config file
CONFIG_PATH = "./"
# Function to load yaml configuration file
def load_config(config_name):
with open(os.path.join(CONFIG_PATH, config_name)) as file:
config = yaml.safe_load(file)
return config
config = load_config("Config.yaml")
# split data into train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=config["test_size"], random_state=config["random_state"]
)
Here are the changes we made:
- The test_size = 0.2 is replaced with config[“test_size”]
- The random state = 42 is replaced with config[“random_state”]
After making similar changes across, the final file would look like this.
# Import important packages
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
import joblib
import os
import yaml
# folder to load config file
CONFIG_PATH = "./"
# Function to load yaml configuration file
def load_config(config_name):
with open(os.path.join(CONFIG_PATH, config_name)) as file:
config = yaml.safe_load(file)
return config
config = load_config("Config.yaml")
# load data
data = pd.read_csv(os.path.join(config["data_directory"], config["data_name"]))
# replace "?" with -99999
data = data.replace("?", -99999)
# drop id column
data = data.drop(config["drop_columns"], axis=1)
# Define X (independent variables) and y (target variable)
X = np.array(data.drop(config["target_name"], 1))
y = np.array(data[config["target_name"]])
# split data into train and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=config["test_size"], random_state= config["random_state"]
)
# call our classifer and fit to our data
classifier = KNeighborsClassifier(
n_neighbors=config["n_neighbors"],
weights=config["weights"],
algorithm=config["algorithm"],
leaf_size=config["leaf_size"],
p=config["p"],
metric=config["metric"],
n_jobs=config["n_jobs"],
)
# training the classifier
classifier.fit(X_train, y_train)
# test our classifier
result = classifier.score(X_test, y_test)
print("Accuracy score is. {:.1f}".format(result))
# save our classifier in the model directory
joblib.dump(classifier, os.path.join(config["model_directory"], config["model_name"]))
You can find the entire code on Github.
So far, we have successfully built a classification model, built a YAML config file, loaded the config file on Jupyter notebook, and parameterized our entire code. Now, if you make changes to the config file and run the notebook.ipynb, you will see the model results very similar to the conventional approach.
We will move to the last section where we will generate a report of everything we have done so far.
Generating reports:
Here are the steps to be followed to generate the report:
- Open the terminal as administrator and navigate to your project folder.
- We will be using nbconvert library for report generation. If it is not installed then do a pip install nbconvert or conda install nbconvert
-
Type jupyter nbconvert –execute –to html notebook.ipynb in the terminal. The –execute executes all the cells in the Jupyter notebook.
-
A notebook.html file will be generated and saved in your project folder.
If you wish to experiment on your model then instead of making changes in your code directly, make changes to your Config.yaml and follow the above steps to generate the report.
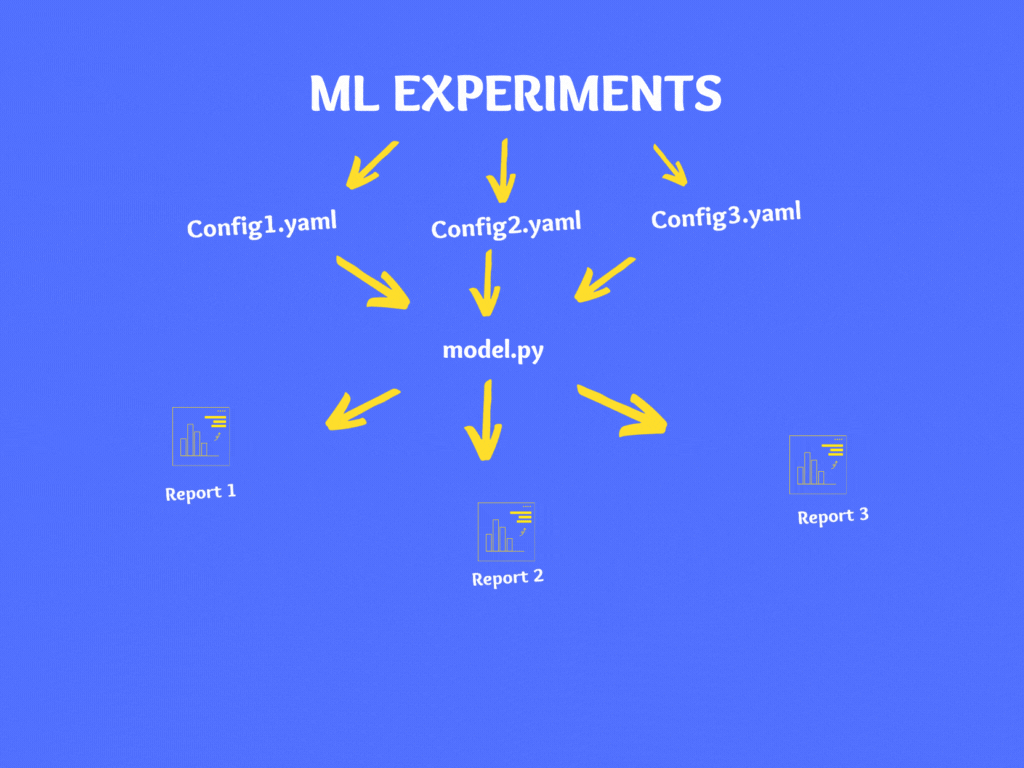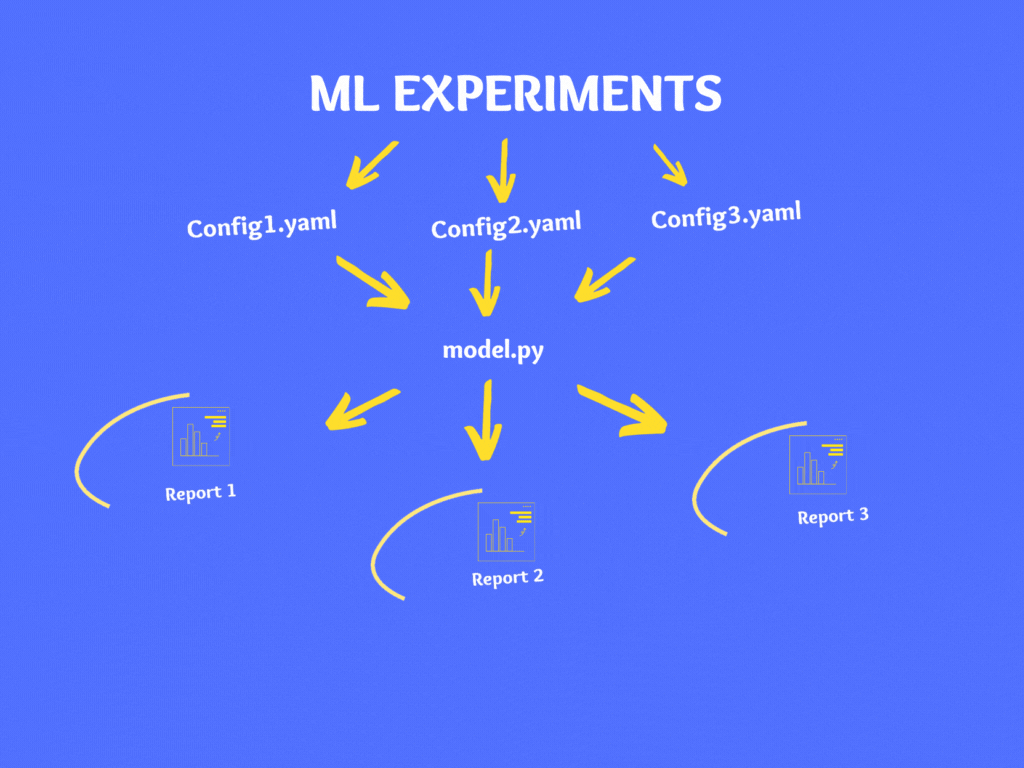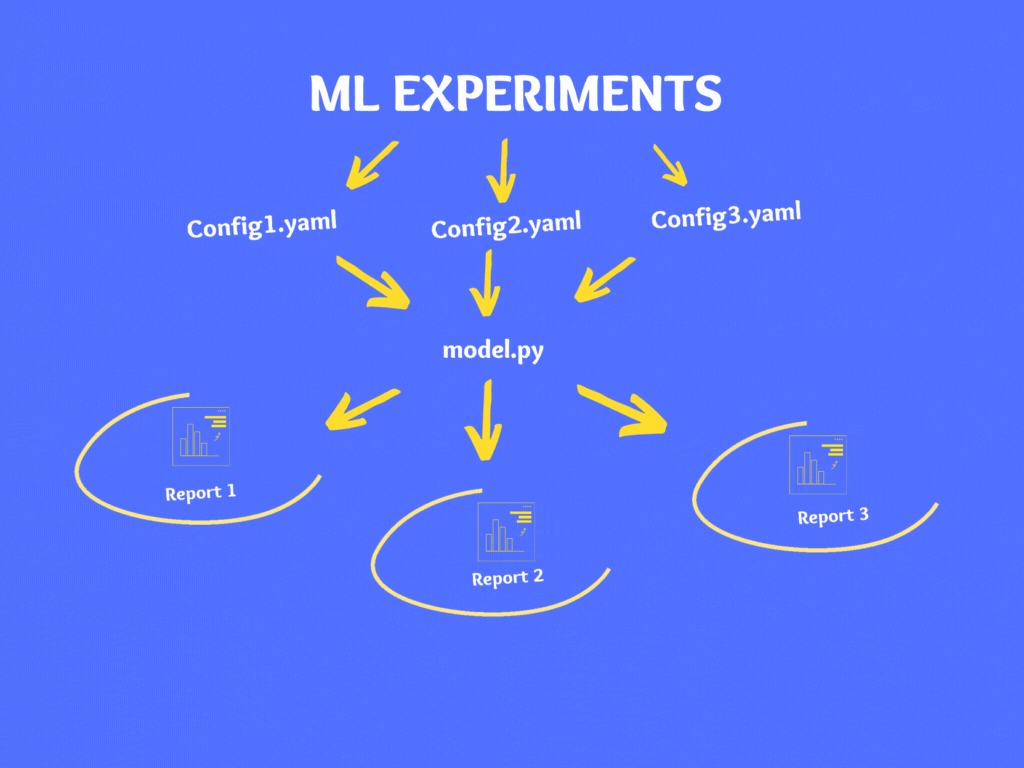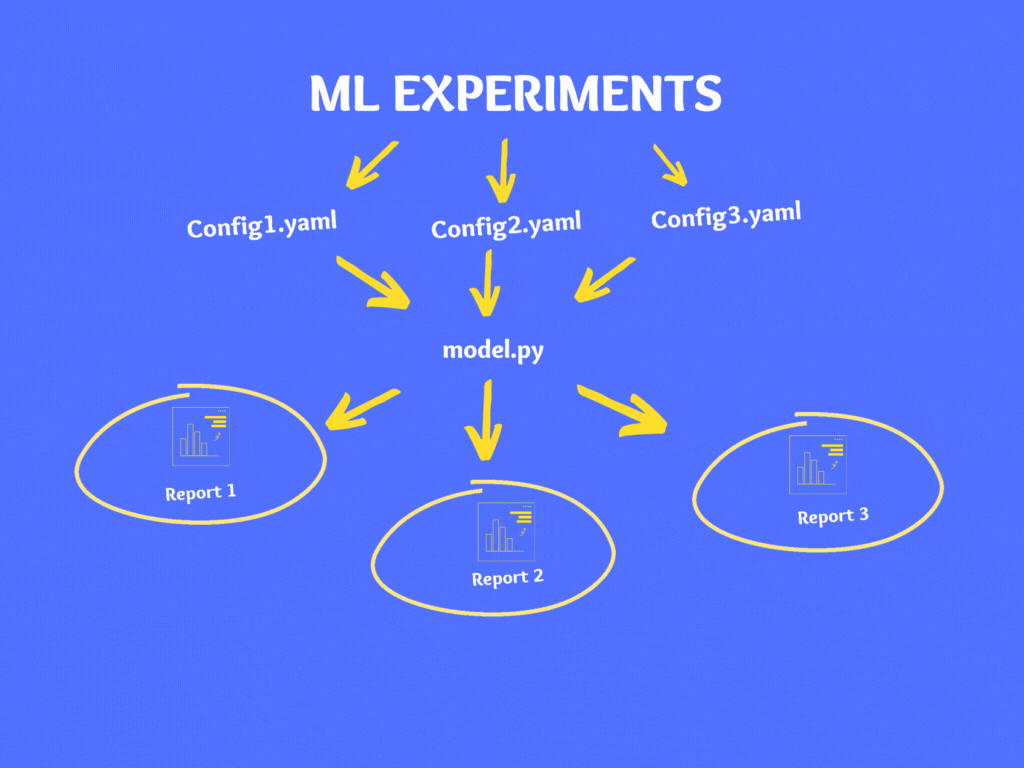
Conclusion:
Now we understand the importance of using a configuration file in a Machine learning project. In this article, we learned what is a configuration file, the importance of the configuration file in your machine learning project, how to create a YAML file and use it in your ML project. Now you can start using the configuration file in your next machine learning project.
Happy learnings !!!!
You can connect with me – Linkedin
You can find the code for reference – Github
References:
https://unsplash.com/
https://yaml.org/
The media shown in this article are not owned by Analytics Vidhya and is used at the Author’s discretion.




