This article was published as a part of the Data Science Blogathon

Learning Objectives
This article provides the following learning objectives-
- Introduction to JupyterLab
- JupyterLab for data scientists
- Advantages of JupyterLab
- Updates in JupyterLab 3.0
- Installation of JupyterLab 3.0
Introduction
The Jupyter community has exponentially grown over the years due to its intuitive data science features. An advanced jupyter project has been released, known as JupyterLab which is one step ahead of Jupyter notebooks.
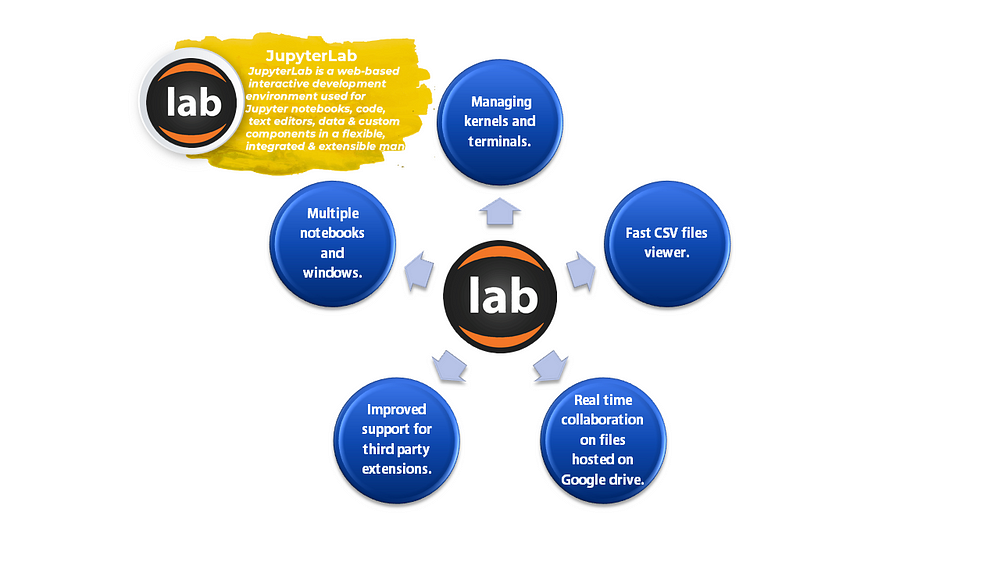
JupyterLab is an open-source interactive web-based user interface for Project Jupyter. It is a development environment for working with notebooks, code, and data. JupyterLab has full support for Jupyter notebooks with multiple integration facilities. JupyterLab enables the usage of text editors, terminals, debuggers, data file viewers simultaneously in a tabbed workspace. It supports multiple notebooks as tabs in the same window. JupyterLab is famous for its Jupyter notebook-based IDE.
Why use JupyterLab for data science?
There are more than one reasons to use JupyterLab for data science. JupyterLab has been primarily developed as an optimal platform for streamlining end-to-end data science pipelines. It has become an indispensable tool for data scientists all over the world.
Exploratory Data Analysis
JupyterLab provides extraordinary features for displaying independent in-line visualizations. Many times there is a requirement of viewing a particular output. This use case can be solved easily by using the JupyterLab 3.0 interface unlike any other traditional IDE like VSCode. In Jupyter notebook, any cell can be executed at any time in the notebook to produce an output. This can be extremely useful for performing EDA. JupyterLab lets users display visualizations and markdowns alongside the code itself enabling a better representation and explanation.
Caching
While working on complex data science problems it is an additional advantage to keep track of the state of execution of each cell. Jupiter’s automatic caching allows storage of the output of every cell for future reference.
Language Independence
JupyterLab provides platform and language independence. It has support for most of the common languages used by data scientists such as Python, R, and Julia along with numerous file formats like Html, markdown, etc.
Interactive IDE
Jupyter Notebook provides standard user interfaces for exploring code and data interactivity. Allowing the users to edit and run the code on the way thus making the code dynamic. Interactiveness improves code performance and makes way for users to make changes according to their needs.
Advantages of JupyterLab 3.0
It’s time to switch to JupyterLab 3.0 which has all the latest upgrades along with all the exciting jupyter features. There’s not one, but multiple reasons for using JupyterLab 3.0.
Unified platform
JupyterLab 3.0 provides a single environment for notebooks, editors, directory discovery, and debuggers. We can work with different file formats such as jpeg, pdf, CSV, etc.
Workspace flexibility
We can arrange files and activities side by side by using tabs and splitters under the same roof. JupyterLab provides drag and drops features to simultaneously perform various activities in a tabbed environment. We can easily resize the tabs for better usage. Right, and left Sidebars panels are available for workspace efficiency.
Code Console
Saving the code computation history can serve great advantage in many ways. They can be used as a record of the code in the notebooks saved in the console. Many times we need to validate our code block where the code console can be handy.
Multiple views
Highlighted views of the same notebook can be presented in several ways for a better coding experience. A particular part of the notebook can be viewed separately side-by-side for comparison inside the JupyterLab workspace. Any made in the view will be reflected and saved in the notebook. This helps to view the necessary information without the hassle of scrolling back and forth.
Copying Cells between multiple notebooks
With jupyter notebooks, we could shift and copy cells within the same notebook. But with JupyterLab we can copy cells between multiple notebooks by simple drag and drop facility.
https://cluebots.com/blog/default-anaconda-applications
Updates in the jupyterLab 3.0
JupyterLab has been regularly coming up with newer updates to enhance the user experience with each version. JupyterLab 3.0 has also introduced some amazing functionalities-
Visual debugger
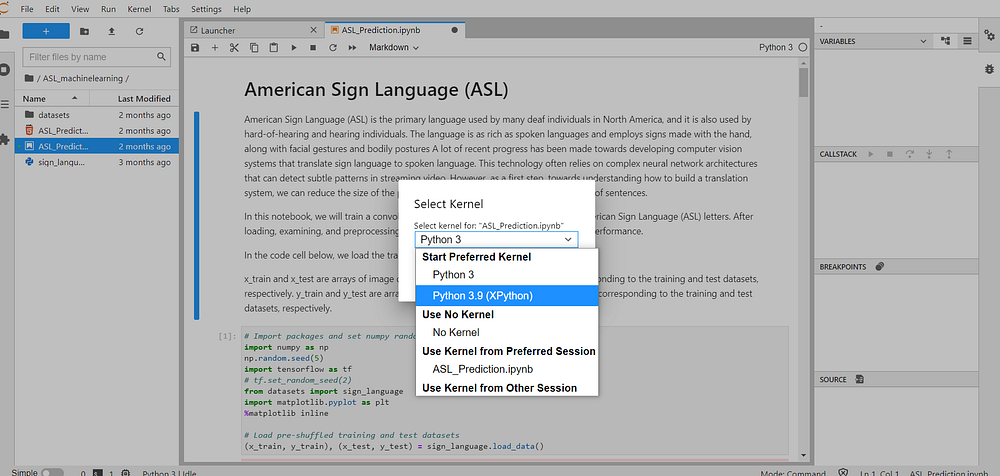
A default front-end debugger has been introduced that enables notebooks, code consoles, and files to be debugged directly from the JupyterLab interface!
The visual debugger can be made available, by installing a kernel with debugging support. The following kernels support the Jupyter Debug Protocol:
- xeus-python: Jupyter kernel for the Python programming language also called xpython.
- xeus-robot: Jupyter kernel for Robot Framework
Installation of xeus-python
First, create a new conda environment then start with the installation.
conda create -n newenv -c conda-forge jupyterlab=3 xeus-python conda activate newenv
Now the xeus-python kernel can be selected kernel from the launcher or using the kernel selection dialogue.

Multiple languages support
Why limit the interface content to a single language when you can select any language of your choice?
JupyterLab3.0 provides the facility to change your interface language according to your choice by installing a language pack. This link lists the language packs available.
Installation of the language pack
The code can be changed according to the language <language code>
conda install -c conda-forge jupyterlab-language-pack-<language code>
After installation, language settings need to be reset in the interface to activate the language. Accept the confirmation box and refresh to view the changed interface.
Extensions installation without NodeJS binding
Extensions are a crucial part of JupiterLab interactive experience, as JupyterLab is formed over an extensible system.
Another way of distributing and installing extensions has been made available in JupyterLab 3.0 as pip or Conda packages. Earlier versions required NodeJS binding with JupyterLab to Install extensions. Now the hassle of rebuilding JupyterLab and the distribution of extensions as npm packages can be avoided by directly installing.
conda install -c conda-forge jupyterlab-python-file
OR
pip install jupyterlab-python-file
The installed extension is a python file, it can be replaced with any JupyterLab extension.
Interface changes
User-side interface changes include-
- Sidebar panels interacting with a specific document have shifted to the right side
- The command palette now comes floating on the center of the top of the window. This enables users to quickly invoke a command while keeping the sidebar closed or switching sidebar panels. You can access the command palette using the View menu or directly using the shortcut Ctrl+Shift+C.
- The file browser now has a filter input feature that allows filtering the list of files using the same fuzzy matching as the command palette.
- The Simple Interface mode has been updated to view the current file in use in the URL bar, as the Jupyter Notebook. We can switch to the Simple Interface mode by a toggle switch, choosing Single Interface mode from the View menu or pressing Ctrl+Shift+D
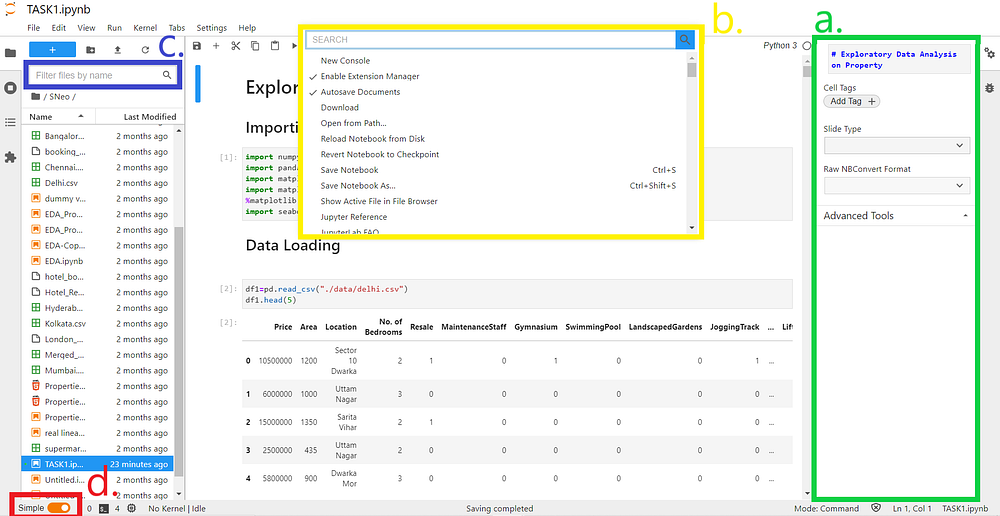 a. Right interactive Sidebar panels
a. Right interactive Sidebar panels
b. Floating command palette
c. filtering files by fuzzy matching
d. Simple Interface mode toggle
Jupyter Server
JupyterLab 3.0 now depends on the Jupyter Server, a new Jupyter project based on the server portion of the classic Notebook server. Jupyter Server is the backend for the core services needed for Jupyter web applications.
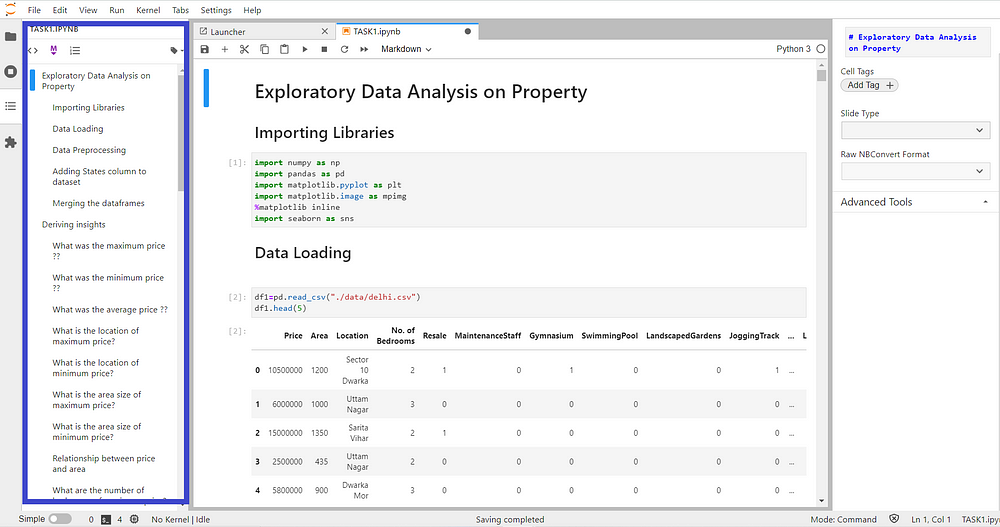
Table of Contents in core
Table of content extension as a part of core provides an easy way to view and navigate through the notebooks and files. We can view the table of content by clicking on the auto-numbering Toggle along with it the description can also be seen. This extension is one of the most used extensions and provides a more user-rich experience.
Installation of JupyterLab 3.0
JupyterLab can be installed using various means such as-
- conda
- mamba
- pip
- pipenv
- docker
Via Conda
conda install -c conda-forge jupyterlab
Via mamba
mamba install -c conda-forge jupyterlab
Via pip
pip install jupyterlab
Via pipenv
pipenv install jupyterlab pipenv shell
virtualenv should be activated if using pipenv to launch jupyter lab.
Via docker
JupyterLab3.0 can be installed by selecting any ready-to-run Docker images maintained by the Jupyter Team. The docker command should include
-e JUPYTER_ENABLE_LAB=yesto enable jupyterLab inside the container.
Other than this Official documentation should be referred for further information about JupyterLab 3.0.
The media shown in this article are not owned by Analytics Vidhya and are used at the Author’s discretion.




