Introduction
When working on a machine learning project, you need to follow a series of steps until you reach your goal, one of the steps you have to execute is hyperparameter optimization on your selected model. This task always comes after the model selection process (select the best model that is performing well than other models).
What is hyperparameter optimization?
Before I define hyperparameter optimization you need to understand what is a hyperparameter. In short, hyperparameters are different parameter values that are used to control the learning process and have a significant effect on the performance of machine learning models. Example of hyperparameters in the Random Forest algorithm is the number of estimators (n_estimators), maximum depth(max_depth), and criterion. These parameters are tunable and can directly affect how well model trains.
Then hyperparameter optimization is a process of finding the right combination of hyperparameter values in order to achieve maximum performance on the data in a reasonable amount of time. It plays a vital role in the prediction accuracy of a machine learning algorithm. Therefore Hyperparameter optimization is considered the trickiest part of building machine learning models.
Most of these machine learning algorithms come with the default hyperparameters values. The default values do not always perform well on a different type of Machine Learning projects you have, that why you need to optimize them in order to get the right combination that will give you the best performance.
A good choice of hyperparameters can really make an algorithm shine.
There are some common strategies for optimizing hyperparameters:
(a) Grid Search
This is a widely used traditional method that performing hyperparameter tuning in order to determine the optimal values for a given model. The Grid search works by trying every possible combination of parameters you want to try in your model, this means it will take a lot of time to perform the entire search which can get very computationally expensive.
NB: You can learn how to implement Grid Search here.
(b) Random Search
This method works differently where random combinations of the values of the hyperparameters are used to find the best solution for the built model. The drawback of Random Search is sometimes could miss important points(values) in the search space.
NB: You can learn more to implement Random Search here.
Alternative Hyperparameter Optimization techniques.
In this series of articles, I will introduce to you different alternative advanced hyperparameter optimization techniques/methods that can help you to obtain the best parameters for a given model. We will look at the following techniques.
- Hyperopt
- Scikit Optimize
- Optuna
In this article, I will focus on the implementation of Hyperopt.
If you’re not performing hyperparameter optimization, you need to start now.
What is Hyperopt
Hyperopt is a powerful python library for hyperparameter optimization developed by James Bergstra. Hyperopt uses a form of Bayesian optimization for parameter tuning that allows you to get the best parameters for a given model. It can optimize a model with hundreds of parameters on a large scale.
Features of Hyperopt
Hyperopt contains 4 important features you need to know in order to run your first optimization.
(a) Search Space
The hyperopt have different functions to specify ranges for input parameters, these are stochastic search spaces. The most common options for a search space to choose are :
- hp.choice(label, options) — This can be used for categorical parameters, it returns one of the options, which should be a list or tuple.Example: hp.choice(“criterion”, [“gini”,”entropy”,])
- hp.randint(label, upper) — This can be used for Integer parameters, it returns a random integer in the range (0, upper).Example: hp.randint(“max_features”,50)
- hp.uniform(label, low, high) — It returns a value uniformly between
lowandhighExample: hp.uniform(“max_leaf_nodes”,1,10)
Other option you can use are:
- hp.normal(label, mu, sigma) — This returns a real value that’s normally-distributed with mean mu and standard deviation sigma
- hp.qnormal(label, mu, sigma, q) — This returns a value like round(normal(mu, sigma) / q) * q
- hp.lognormal(label, mu, sigma) — This returns a value drawn according to exp(normal(mu, sigma))
- hp.qlognormal(label, mu, sigma, q) — This returns a value like round(exp(normal(mu, sigma)) / q) * q
You can learn more search space options here.
NB: Every optimizable stochastic expression has a label (e.g n_estimators) as the first argument. These labels are used to return parameter choices to the caller during the optimization process.
(b) Objective Function
This is a function to minimize that receives hyperparameters values as input from the search space and returns the loss. This means during the optimization process, we train the model with selected hyperparameters values and predict the target feature and then evaluate the prediction error and give it back to the optimizer. The optimizer will decide which values to check and iterate again. You will learn how to create an objective function in a practical example.
(c) fmin
The fmin function is the optimization function that iterates on different sets of algorithms and their hyperparameters and then minimizes the objective function. the fmin takes 5 inputs which are:-
- The objective function to minimize
- The defined search space
- The search algorithm to use such as Random search, TPE (Tree Parzen Estimators), and Adaptive TPE.
NB:hyperopt.rand.suggestandhyperopt.tpe.suggestprovides logic for a sequential search of the hyperparameter space. - The maximum number of evaluations.
- The trials object (optional).
Example:
fPython Code:
(d) Trial Object
The Trials object is used to keep All hyperparameters, loss, and other information, this means you can access them after running optimization. Also, trials can help you to save important information and later load and then resume the optimization process. (you will learn more in the practical example).
from hyperopt import Trials trials = Trials()
After understanding the important features of Hyperopt, the way to use Hyperopt is described in the following steps.
- Initialize the space over which to search.
- Define the objective function.
- Select the search algorithm to use.
- Run hyperopt function.
- Analyze the evaluation outputs stored in the trials object.
Hyperpot in Practice
Now that you know the important features of Hyperopt, in this practical example, we will use Mobile Price Dataset and the task is to create a model that will predict how high the price of the mobile is 0(low cost) or 1(medium cost) or 2(high cost) or 3(very high cost).
Install Hyperopt
You can install hyperopt from PyPI.
pip install hyperopt
Then import important packages include Hyperopt.
# import packages
import numpy as np
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
from sklearn.model_selection import cross_val_score
from sklearn.preprocessing import StandardScaler
from hyperopt import tpe, hp, fmin, STATUS_OK,Trials
from hyperopt.pyll.base import scope
import warnings
warnings.filterwarnings("ignore")
Dataset
Let’s load the dataset from the data directory. To get more information about the dataset read here.
# load data
data = pd.read_csv("data/mobile_price_data.csv")
Check the first five rows of the dataset.
#read data data.head()
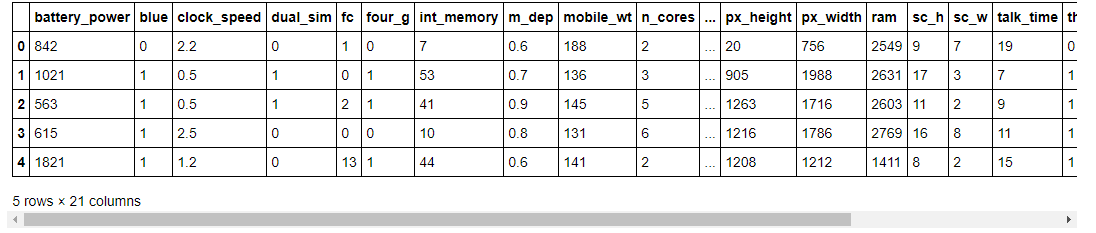
As you can see, in our dataset we have different features with numerical values.
Let’s observe the shape of the dataset.
#show shape data.shape
(2000, 21)
In this dataset, we have 2000 rows and 21 columns. Now let’s understand the list of features we have in this dataset.
#show list of columns list(data.columns)
[‘battery_power’, ‘blue’, ‘clock_speed’, ‘dual_sim’, ‘fc’, ‘four_g’, ‘int_memory’, ‘m_dep’, ‘mobile_wt’, ‘n_cores’, ‘pc’, ‘px_height’, ‘px_width’, ‘ram’, ‘sc_h’, ‘sc_w’, ‘talk_time’, ‘three_g’, ‘touch_screen’, ‘wifi’, ‘price_range’]
You can find the meaning of each column name here.
Splitting the dataset into Target feature and Independent features
This is a classification problem, we will then split the target feature and independent features from the dataset. Our target feature is price_range.
# split data into features and target
X = data.drop("price_range", axis=1).values
y = data.price_range.values
Preprocessing the Dataset.
Then standardize the independent features by using the StandardScaler method from scikit-learn.
# standardize the feature variables scaler = StandardScaler() X_scaled = scaler.fit_transform(X)
Define Parameter Space for Optimization
We will use three hyperparameters of the Random Forest algorithm which are n_estimators,max_depth, and criterion.
space = {
"n_estimators": hp.choice("n_estimators", [100, 200, 300, 400,500,600]),
"max_depth": hp.quniform("max_depth", 1, 15,1),
"criterion": hp.choice("criterion", ["gini", "entropy"]),
}
We have set different values in the above-selected hyperparameters. Then we will define the objective function.
Defining a Function to Minimize (Objective Function)
Our function to minimize is called hyperparamter_tuning and the classification algorithm to optimize its hyperparameter is Random Forest. I use cross-validation to avoid overfitting and then the function will return a loss values and its status.
# define objective function
def hyperparameter_tuning(params):
clf = RandomForestClassifier(**params,n_jobs=-1)
acc = cross_val_score(clf, X_scaled, y,scoring="accuracy").mean()
return {"loss": -acc, "status": STATUS_OK}
NB: Remember that hyperopic minimizes the function, that why I add a negative sign in the acc :
Fine Tune the Model
Finally first instantiate the Trial object, fine-tuning the model, and then print the best loss with its hyperparameters values.
# Initialize trials object
trials = Trials()
best = fmin(
fn=hyperparameter_tuning,
space = space,
algo=tpe.suggest,
max_evals=100,
trials=trials
)
print("Best: {}".format(best))
100%|█████████████████████████████████████████████████████████| 100/100 [10:30<00:00, 6.30s/trial, best loss: -0.8915] Best: {‘criterion’: 1, ‘max_depth’: 11.0, ‘n_estimators’: 2}.
After performing hyperparameter optimization, the loss is -0.8915 means the model performance has an accuracy of 89.15% by using n_estimators = 300,max_depth = 11, and criterion = “entropy” in the Random Forest classifier.
Analyze results by using trials object
The trial object can help us to inspect all of the return values that were calculated during the experiment.
(a) trials.results
This shows a list of dictionaries returned by ‘objective’ during the search.
trials.results
[{‘loss’: -0.8790000000000001, ‘status’: ‘ok’}, {‘loss’: -0.877, ‘status’: ‘ok’}, {‘loss’: -0.768, ‘status’: ‘ok’}, {‘loss’: -0.8205, ‘status’: ‘ok’}, {‘loss’: -0.8720000000000001, ‘status’: ‘ok’}, {‘loss’: -0.883, ‘status’: ‘ok’}, {‘loss’: -0.8554999999999999, ‘status’: ‘ok’}, {‘loss’: -0.8789999999999999, ‘status’: ‘ok’}, {‘loss’: -0.595, ‘status’: ‘ok’},…….]
(b) trials.losses()
This shows a list of losses (float for each ‘ok’ trial).
trials.losses()
[-0.8790000000000001, -0.877, -0.768, -0.8205, -0.8720000000000001, -0.883, -0.8554999999999999, -0.8789999999999999, -0.595, -0.8765000000000001, -0.877, ………]
(c) trials.statuses()
This shows a list of status strings.
trials.statuses()
[‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ‘ok’, ……….]
NB: This trial object can be saved, passed on to the built-in plotting routines, or analyzed with your own custom code.
Wrapping Up
Congratulations, you have made it to the end of this article!
You can download the dataset and notebooks used in this article here:
https://github.com/Davisy/Hyperparameter-Optimization-Techniques
About the Author
Davis David
Davis David is the Data scientist & software developer with a background in Computer Science. He is passionate about Artificial Intelligence, Machine Learning, Deep Learning, and Big Data. Davis is the Co-Organizer and Facilitator of the Artificial Intelligence (AI) movement in Tanzania by conducting AI meetups, workshops, and events with a passion to build a community of Data Scientists in Tanzania to solve our local problems. Also, he organized Pythontz and TanzanAi Lab Community and became Ambassador of ZindiAfrica in Tanzania.




